Received 27 November 2020; Revised 14 December 2020; Accepted 15 December 2020; Published 29 December 2020
Ramadan M. Ramadan, Rania G. Mohamed, Mona Hassan, and Samir M. El-Medani, Spectroscopic and Theoretical Studies of Some Bivalent Metal Complexes of a Quinoline Schiff Base Derivative, Journal of Transition Metal Complexes, 3 (2020), art246114. doi:10.32371/jtmc/246114
Interaction of M(II) ions, M=Co, Ni, Cu, Zn, and Pd, with a Schiff base ligand (HL), derived from the condensation of 2-hydroxy-5-nitrobenzaldehyde and 8-aminoquinoline, gave the complexes [Co(L)2(H2O)2], [Ni(L)2(H2O)2], [Cu(L)(H2O)]CH3COO, [Zn(L)(H2O)]CH3COO, and [Pd(HL)Cl2]. The ligand and its complexes were characterized by means of elemental and thermal analyses, molar conductance, and magnetic moment measurements along with the different spectroscopic techniques. Theoretical calculations based on DFT were carried out to verify the optimized structures of ligand and complexes. The relative reactivity of the compounds was estimated from chemical descriptors analysis using the HOMO and LUMO frontier orbitals. Molecular docking of ligand and its zinc complex revealed that the interaction of Zn(L) with DNA is comparable to that of HL due to the presence of water OH groups instead of the hydroxyl group of ligand.
copper complexes; X-ray analysis; DFT studies; antioxidant activity; CT-DNA binding; molecular docking
In the last few decades, Schiff base derivatives showed a vital importance in the chemistry of organic and coordination compounds [1,2,3,4]. These compounds stemmed their significance from their applications as models in biology, medicinal, and bioinorganic chemistry [5,6], as well as their uses in industrial catalysis, corrosion, and photochromism
[7,8,9]. In addition, photo- and thermochromic properties of the Schiff bases and their metal complexes make them significant in modern technology. They are found to be valuable in optical computers to measure and control of the radiation of imaging systems, in the molecular memory storages and photo detectors in biological systems [10]. Moreover, heterocyclic Schiff bases containing one or more nitrogen atoms are considered as good ligands. This is due to the presence of nitrogen atoms in the rings with a localized pair of electrons [11,12,13]. In particular, the quinoline framework containing Schiff base derivatives were found to serve as important heterocyclic compounds
with diverse array of biological properties, such as antiasthmatic, anti-bacterial, and anti-inflammatory agents [14,15]. Moreover, the quinolines occurred as based natural products as quinine and chloroquine, which have been extensively used as drugs for the treatment of malaria [16]. Also, the chloro-aminoquinoline Schiff base derivatives were screened for their anticholinesterase activity, which assessed for Alzheimer disease [17]. The inhibitory activity of these compounds and their copper derivatives was
evaluated by Ellman’s spectrophotometric method in acetylcholinesterase (ACHE) and butyrylcholinesterase (BCHE) from equine serum. Docking and molecular dynamics studies of ACHE were performed and the results suggested that these compounds are able to bind inside ACHE similarly to other inhibitors [17]. On the other hand, many studies of mixed ligand complexes with nitrogen-containing heterocyclic derivatives (such as 8-hydroxyquinoline and 2-aminothiazole) and the corresponding Schiff base ligands were reported. These derivatives have interesting structural
features and in vitro antimicrobial and cytotoxicity activities [18]. It was found that the presence of aromatic and/or heteroaromatic groups in the structure of nitrogen gives these ligands additional properties and therapeutic, pharmacologic, and DNA cleavage activity [19,20].
On continuation of our interest in synthesis and investigations of many Schiff base ligands and their transition metal complexes [2,4,21,22,23], here we report the synthesis of a Schiff base derived from the condensation of 2-hydroy-5-nitro benzaldehyde and 8-aminoquinoline as well as its Co(II), Ni(II), Cu(II), Zn(II), and Pd(II) complexes. Different spectroscopic tools, biological activities along with DFT calculations and molecular docking were used to characterize the ligand and the complexes.
CoCl2·6H2O, Ni(CH3COO)2·4H2O, Cu(CH3COO)2·H2O, Zn(CH3COO)2·2H2O, and dichloro-1,5-cyclooctadiene-palladium, 2-hydroy-5-nitro benzaldehyde and 8-aminoquinoline were purchased from Aldrich. All solvents were of analytical grade and purified by distillation.
IR spectra (KBr pellets) were recorded on a Unicam-Mattson 1000 FT-IR spectrometer. NMR measurements were performed on a Varian Mercury VX-300 NMR spectrometer. Samples were dissolved in DMSO-d6. Magnetic susceptibilities of the paramagnetic complexes in the solid state (Gouy method) were measured on a Sherwood Scientific magnetic susceptibility balance. Conductivity measurements were carried out in DMSO (1 × 10−3 M) at 25 °C using Jenway 4010 conductivity meter. Mass spectra of the solid complexes (70 eV, EI) were performed on a Finnigan MAT SSQ 7000 spectrometer. Bruker EMX spectrometer was used to record the electron spin resonance spectra at 25 °C in the X-band frequency 9.714 GHz, and on a microcrystalline powder with microwave power around 2.012 mW. Thermogravimetric analyses were carried out under nitrogen atmosphere with a heating rate of 10 °C/min using a Schimadzu DT-50 thermal analyzer. Elemental analyses were performed on a Perkin-Elmer 2400 CHN elemental analyzer.
The Schiff base ligand was synthesized by refluxing a mixture of 2-hydroy-5-nitro benzaldehyde (1.67 g, 10 mmol) and 8-aminoquinoline (1.44 g, 10 mmol) in 25 mL absolute ethanol for 3 h. The reaction mixture was cooled and the solvent was evaporated. The residue obtained was washed using cold ethanol, and then recrystallized from hot
ethanol. A crystalline orange product was left to dry for few hours (yield 83%), Scheme 1.
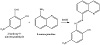
Scheme 1: Synthesis of HL.
A general procedure was employed to synthesize the reported metal complexes. A 1:1 molar ratio from the ligand and metal salt was used except for the cobalt and nickel complexes (1:2 metal to ligand). The reaction mixture in methanol was refluxed for 3 h until the color was changed. The solution was left to stand overnight. The residue was separated by filtration, washed with hot petroleum ether, and then recrystallized from hot ethanol. The fine crystals formed were left to dry under vacuum for several hours. Table 1 gives the color, yield, elemental analyses, and mass spectral data of the reported compounds.

Table 1: Color, yield, elemental analyses, and mass spectra data for the reported compounds.
In vitro antibacterial and antifungal activity of the ligand and its complexes were tested against four bacteria: Escherchia coli and Enterobacter aerogenes as Gram-negative bacteria and Staphylococcus aureus and Enterococcus faecalis as Gram-positive bacteria, and the fungus: Aspergillus niger. The tests were carried out using paper disc diffusion method (The Central lab, Cairo University, Egypt). The detailed procedure was previously reported [2,21]. The obtained data for antibacterial were compared with those of doxycycline as an antibacterial standard.
All the calculations were performed using the hybrid density functional theory (DFT) method B3LYP as implemented in the Gaussian 09 software package [24]. The geometries were optimized using the standard double zeta plus polarization basis set 6-31G (d,p) for the ligand atoms and effective core potential basis set LANL2DZ for the metal complexes. The purpose of the quantum mechanics calculations validates the proposed 3D structure of the obtained compounds as well as to find out key factors for their reactivity. The theoretical calculations of the spectral modes of vibrations of ligand were carried out using DFT.
The docking studies were performed using AutoDock Vina 4 [25]. Two macromolecule targets (PDB ID: 4MZI and 1ZEW) were used for docking the ligand and its zinc complex. 4MZI is a p53 tumor suppressor protein, which is the most frequently mutated gene in human cancer [26]. 1ZEW is an X-ray crystal structure of a B-DNA of inverted repeat DNA sequence d(CCnnnN6N7N8GG), where N6, N7, and N8 are naturally occurring nucleotides [27]. The molecular visualization system PyMOL (2.0.7) was used to visualize the docking molecules [28].
Interaction of the quinoline derivative Schiff base ligand HL (Scheme 1) with the bivalent cobalt and nickel ions resulted in the formation of neutral octahedral complexes with the molecular formula [M(L)2(H2O)2]. On the other hand, reactions of Cu2+ and Zn2+ ions with HL gave the two cationic complexes [M(L)(H2O)]CH3COO. In case of the reaction
of PdCl2 with HL, the neutral complex [Pd(HL)Cl2] was formed without the loss of ligand proton. The ligand and its metal complexes were characterized using different spectroscopic techniques (IR, 1H NMR, mass), elemental analyses, magnetic measurements, molar conductance, and thermal analysis. Elemental analyses and mass spectra data of the reported compounds were in accordance with the proposed molecular formulas. The molar conductivities of 1 × 10−3 M
solutions of the complexes at 25 °C were found to be in the range 11–16 Ω−1 mol−1 cm2 (Table 1) indicating nonor weak electrolyte characteristics of these derivatives. The magnetic susceptibility measurements for the cobalt, nickel, and copper paramagnetic complexes at 298 K gave effective magnetic moment (μeff) values in accordance with the corresponding unpaired electrons, Table 1. The observed small deviation from the spin-only moments could be due to the spin-orbit coupling.
The IR spectra (KBr pellets) of the ligand and its complexes were recorded in the region 4000–400 cm−1. The most prominent IR and 1H NMR spectral data for HL and complexes are given in Tables 2 and 3. The experimental and calculated vibrational modes of HL were investigated to assign and compare the modes of the important functional groups such as ν(OH), ν(C=N), and ν(NO2) [2]. The IR spectrum of HL displayed a strong and broad band due to νOH. The shift of the band towards lower frequency than those of usual free OH groups could be owing to the existence of hydrogen bond between hydroxyl proton and the azomethine nitrogen. Also, the spectrum exhibited two medium stretching frequency bands at 1,437 cm−1 and 1,238 cm−1 due to asymmetric and symmetric νNO2. The 1H NMR spectrum of HL in deuterated DMSO showed a slightly broad singlet at 10.31 ppm, which is assigned to the hydroxyl proton. This signal disappeared on addition of D2O to solution. The spectrum also displayed a singlet due to the azomethine proton and two multiples for the quinoline and phenyl rings, Table 2. The IR spectra of the reported metal complexes showed the functional groups of the ligand, such as C=N and NO2 groups, with the appropriate shift due to complex formation, Table 3. Interestingly, except for the palladium complex, the IR spectra of the other complexes showed the disappearance of OH band. It indicated that the ligands coordinated to metal from the hydroxyl oxygen and the azomethine nitrogen [2,21,22,23]. The shifts in the bands of the other functional groups, towards low frequencies, also confirmed such a conclusion. Furthermore, the IR spectra of the complexes displayed new nonligand bands corresponding to stretching frequencies of the M−O and M−N moieties [29]. The 1H NMR spectra for the zinc and palladium complexes gave more insight to their structures. It is worth to mention that the NMR spectrum of the palladium complex displayed a broad singlet at 12.20 ppm for the hydroxyl hydrogen. This signal was shifted to lower energy relative to that of ligand (10.31 ppm). Scheme 2 gives the proposed structures of the isolated complexes.

Table 2: Assignments of the important experimental and calculated vibrational modes and 1H NMR data of HL ligand.

Table 3: IR and NMR data for the reported complexes.
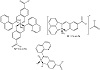
Scheme 2: The proposed structures of complexes.
Electron spin resonance (ESR) is a powerful tool for investigating the structure and dynamics of molecular systems containing paramagnetic center. The ESR spectrum of the microcrystalline powder copper complex, [Cu(L)(H2O)]CH3COO, was recorded at room temperature on the Klystron X-band at frequency 9.714 GHz, and with microwave power around 2.012 mW and the g factors were measured relative to the standard indicator DPPH (g = 2.0037); see Figure 1. The ESR signals in parallel and perpendicular regions appeared as partially resolved signal due to either distortion in geometry or electron-spin-nuclearspin coupling. The spectrum of the complex displayed a g∥ value = 2.065, while the g⊥ value = 2.095. According to Kivelson and Neiman, the g∥ value in the Cu(II) complex can be used as a measure of covalent characteristics of the metal-ligand bond [30]. For the ionic case, the g∥ value is generally ≥ 2.3, while for covalent state it is < 2.3. Since the value of g∥ of the complex is smaller than 2.3, the Cu(II)-ligand bonds have a considerable covalent property. The observed values for g, where g⊥ > g∥ > 2.003, indicated that the unpaired electron is mainly localized in the dz2 orbital of the copper(II) ion [31]. The geometric factor G is a measure of exchange interaction between the copper centers in the solid compounds. The G factor can be calculated using the expression [32]:
|
|
|
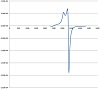
Figure 1: Radiograph of lap joint defects.
If the value of G is larger than 4, the exchange interaction between copper(II) centers is considered negligible. On the other hand, if G is smaller than 4, a significant exchange
interaction can be specified in the complex. The calculated G factors for the present complexes are 0.68 indicating a significant exchange interaction between the copper ions.
The electronic absorption spectrum of HL in either MeOH or CH2Cl2 consists of a UV band (π–π∗) and two broad bands in the visible region, Table 4. The position of the UV band was not changed in the two solvents. The lower wavelength bands correspond to n–π∗ transitions. This showed a little red shift in CH2Cl2 relative to that in methanol. On the other hand, the other visible band (higher wavelength) showed a corresponding red shift in CH2Cl2 solvent with respect to that of methanol one, Table 4. This absorption band could be due to a charge transfer (CT) between HL and the solvent. Obviously, the CT is stronger in case of methanol solvent [33,34]. The spectra of the complexes also displayed two absorption bands for π–π∗ and n–π∗ transitions with the appropriate shift due to complex formation (Table 4). The two complexes, Cu(L) and Zn(L), showed a shoulder to the UV band. Interestingly, the spectra of the complexes did not exhibit CT bands, presumably because the two sites expected to involve the CT (OH and HC=N) were engaged in the coordination.

Table 4: The UV-vis data of HL and its complexes.
In order to give more insight into the structure of the complexes, thermal studies on the solid complexes using TG and differential thermogravimetric (DTG) techniques were performed [35]. The TG and DTG plots of [Co(L)2(H2O)2], Co[C32H24N6O8]; MW = 679.51, exhibited four decomposition steps. The first from 125–350 °C, with a net weight loss of 5.30 (5.25) %, is probably due to elimination of two coordinated H2O; see Table 5. The second step occurred in the temperature range 350–450 with a net weight loss of 22.66%, which is consistent with elimination of (C10H4NO). The third in the temperature range 350–450 with a net weight loss of 10.89% corresponding to elimination of (C3H8NO). The fourth step ranged from 647 °C to 1,000 °C to give finally solid residual of CoO + 4C (Table 5). The TG and DTG plots of [Ni(L)2(H2O)2], Ni[C32H24N6O8]; MW = 679.28, exhibited four decomposition steps in the temperature range 130–1,000 °C. The first step (130–300 °C) showed mass percent of 5.30% which corresponded to elimination of two H2O. The second, third, and fourth steps at temperature ranges 300–426 °C, 426–609 °C, and
609–1,000 °C showed the elimination of mass percent of 24.14%, 17.37%, and 38.57% corresponding to C9H10NO2,
C6H2N2O, and C15H8N3O2 species, respectively, leaving finally NiO + 2C solid species (Table 5). Thermal studies of the copper complex, [Cu(L)(H2O)]CH3COO, were carried out using thermogravimetry. The TG plot of Cu[C18H13N3O5]; MW = 414.868 showed two well defined and nonoverlapping steps in the 216–1,000 °C range (listed in Table 5). The first at 216–477 °C showed a net weight loss of 38.87% corresponding to elimination of C7H6N2O3. The second decomposition peak (477–1,000 °C) showed a net weight loss of 29.95% corresponding to a residual solid of CuO + 5C (Table 5). The TG and DTG plots of [Zn(L)(H2O)]CH3COO, Zn [C18H15N3O6]; MW = 434.723, exhibited three decomposition steps. The first from 120–300 °C, with a net weight loss of 14.72%, is probably due to elimination of one coordinated H2O and CH4NO species; see Table 5. The second step occurred in the temperature range 300–400 with a net weight loss of 4.37%, which is consistent with elimination of (H3O) species. The third in the temperature range 400–1,000 with a net weight loss of 56.59% corresponding to elimination of (C15H6N2O2) to give finally a solid residual of ZnO + 2C (Table 5).

Table 5: Thermogravimetric data of (HL) ligand and its Co(II), Ni(II), Cu(II), Zn(II), and Pd(II) complexes.
The TG and DTG plots of [Pd(HL)Cl2]; Pd[C16H11N3O3Cl2]; MW = 470.58, required three decomposition steps. The first from 50–309 °C, with a net weight loss of 15.09%, is probably due to elimination of Cl2 molecule (Table 5). The second step occurred in the temperature range 309–454 with a net weight loss of 28.69%, consistent with elimination of (C7H9N3). The third in the temperature range 454–1,000 with a net weight loss of 24.22% corresponding to elimination of (C8H2O) to give finally solid residual of PdO2 + C (Table 5).
To investigate the stereochemistry of the most stable structure of the complexes, it was focused first on the structure of the ligand, specifically on the orientation of its functional groups with respect to each other and with respect to the central azomethine (HC=N) part. The stereochemistry and chemical reactivity predictions of the ligand and its complexes were investigated by the hybrid density functional theory (DFT) method at the B3LYP level. The optimized structure of HL (Figure 2) showed that it has a planar arrangement with a C1 point group and a total minimized energy of 16.52 kcal/mol. The dihedral angles N2−C10−C12−C14, C12−C10−C11−H28, and C7−C8−N20−O21 are equal to 180.0 °C, 180.0 °C, and −0.010 °C, respectively, confirming the planarity of the molecule. Also, the angles C9−N2−C10, C5−C9−N2, and O21−N20−O22 have the values 127.53 °C, 119.70 °C, and 123.46 °C, respectively. These values are corresponding to sp2 hybridization and confirming that the molecule is nearly planar. The bond lengths of the azomethine moiety showed that the N2−C9 and N2−C10 bonds have the values 1.31 Å and 1.41 Å, respectively, suggesting double and single bond characteristics. These values are in the normal range observed before [21,36,37]. For example, the X-ray structure analysis of a Schiff base ligand derived from 2-phenylacetohydrazide and 2′-hydroxy acetophenone showed a value of 1.29 Å for the C=N of the azomethine part [21]. The optimized structure for HL showed the presence of hydrogen bonding between N2 and H23 (1.59 Å) to form an O(1)…H(23)…N(2) moiety. This bond separation illustrates the van der Waal displacement between H, the donor (D), and the acceptor (A). The orientation of the functional groups of the HL suggested that it might only coordinate to the metal ion through the nitrogen of the azomethine and the hydroxyl group, that is, it acts as a bidentate ligand with the coordination of NO set of donors.
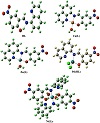
Figure 2: Voids concentration in lap joint versus Ti concentration.
The spectroscopic and analytical investigation of the cobalt and nickel complexes concluded that the metal ion bound to two L moieties and two coordinated water molecules. The energetically stable model of the nickel derivative showed a distorted octahedral geometry with a total minimized energy of 236.42 kcal/mol (Figure 2). The chelated ligand moiety to the Ni ion gave a six-membered ring. The bond lengths for the two Ni−N and the two Ni−O are 1.85 Å, 1.84 Å, 1.79 Å, and 1.80 Å, respectively. Optimized angles for the two N−Ni−O (six-membered cycles) are 92.32° and 85.02°. As shown from the figure, the two coordinated water molecules are attached to the metal with a trans orientation and the O−Ni−O angle is 176.40°. The other bonds and angles of the complex are comparable to those of the ligand itself.
The optimized structures of the two complexes [M(L)(H2O)]+, M=Cu or Zn, were found to be almost identical (Figure 2). The metal existed in a distorted squareplanar environment and the ligand bound to it in a tridentate fashion. The total minimized energy was 187.44 kcal/mol and 190.58 kcal/mol for the copper and zinc derivative, respectively. The dihedral angles of the planes containing the metal ion were C9−C4−N18−Cu33 = 1.74°, C3−N18−Cu33−O29=101.57°, C3−N18−Zn36−N19=176.53°, and C20−O29−Zn36−N18 = 76.65°. In addition, the bond angles between the metal ion and the adjacent atoms were N18−Cu33−N19 = 84.91°, O18−Cu33−O19 =84.91°, O18−Cu33−O29=176.45°, N19−Cu33−O34 = 171.24°, O29−Cu33−O34 = 78.82°, N18−Zn36−N19 = 82.86°, O29−Zn36−O33 = 82.183°, N19−Zn36−O33 = 149.70°, and N19−Zn36−O29 = 93.35°. These data indicated the nonplanarity of the molecules as well as the distorted nature of their square planar arrangements. It can also be noted that in the zinc complex the two nitrogen atoms and the oxygen of the ligand moiety almost lie in the same plane with Zn, while the coordinated water molecule is out of the plane. The bond lengths between copper or zinc and the coordinated donor atoms were in the normal ranges observed before for similar Schiff base complexes [21,38]. The values of bond separation for the copper derivative were found to be N18−Cu33 = 1.99 Å, N19−Cu33 = 1.97 Å, O29−Cu33 = 1.90 Å, and Cu33−O34 = 2.02 Å. On the other hand, the bond lengths for the zinc complex were N18−Zn36 = 2.0655 Å, N19−Zn36 = 2.0455 Å, O29−Zn36 = 1.9399 Å, and O33−Zn36 = 2.0516 Å.
In case of the [Pd(HL)(Cl)2] complex, the optimized structure showed that palladium species existed in a distorted tetrahedral skeleton with two coordinated chlorine atoms, and the ligand coordinated to it in a bidentate mode (Figure 2). The total minimized energy was 157.88 kcal/mol. From the figure, one can notice that the palladium metal and ligand moiety are nearly in the same plane, while the two chlorine atoms are out the plane in opposite positions. The distortion in the tetrahedral arrangement around the metal stemmed from the bond angles and dihedral angles. The calculated dihedral angles for the complex were Cl34−Pd33−Cl35 = 108.23°, C1−N19−Pd33−O29 = 12.44°, C20−O29−Pd33−Cl35 = 41.78° and C9−N19−Pd33−Cl34 = 72.53°. Also, the bond angles N19−Pd33−O29 = 106.64°, N19−Pd33−Cl34 = 112.12°, N19−Pd33−Cl35 = 108.88°, and O29−Pd33−Cl34 = 109.72° obviously deviated from regular tetrahedron. The bond lengths between palladium and nitrogen, oxygen and chlorine atoms were found to be N19−Pd33 = 1.952 Å, O29−Pd33 = 1.922 Å, and Pd33−Cl34 = 2.27 Å. They were in the normal range for these bonds as observed before [39,40]. For example, the palladium complex [PdCl2(L)], L = 2,6-diisopropyl-N-[(S)-pyrrolidin-2-ylmethyl]-aniline, exhibited Pd-N bond lengths equal to 2.04 Å and 2.07 Å, and Pd-Cl bond lengths equal to 2.31 Å and 2.32 Å [39].
The global chemical reactivity parameters including HOMO, LUMO, energy gap (ΔE), electronegativity (X), chemical potential (V), electron affinity (A), ionization potential (I), chemical hardness (η), chemical softness (S), and electrophilicity (ω) of the reported compounds are given in Table 6 [41]. The estimated frontier molecular orbital energies for the compounds are illustrated in Figure 3. The HOMO orbital energy represents electron donating ability, while the LUMO orbital energy characterizes electron withdrawing ability. The energy gap between them shows the molecular chemical stability. This parameter is crucial for determining molecular electrical transport properties. The smaller energy gap reflects the easiness of the CT, and the polarization, which occurs within the molecule [42]. Therefore, the order of increasing reactivity of the reported complexes is Pd(HL) > Zn(L) > Ni(L)2 > Cu(L). The electronegativity parameter (X) is a reflection for the electrostatic potential, where the electron partially transferred from one of lower electronegativity to another of higher electronegativity [43]. The calculated data showed that the order of decreasing X is Cu(L) > Zn(L) > Pd(HL) > Ni(L)2. On the other hand, the results of small chemical hardness values for the reported derivatives reflect the ability of charge-transfer within the molecule. The order of increasing the charge-transfer within the molecule is Pd(HL) < Zn(L) < Ni(L)2 < Cu(L).

Table 6: The global chemical reactivity descriptors for HL and its complexes.

Figure 3: Joint strength (Mpa) versus Ti concentration (wt%).
The in vitro antimicrobial activities of the ligand and its complexes were screened against four bacteria: E. coli and E. aerogenes as Gram-negative bacteria and S. aureus and E. faecalis as Gram-positive bacteria, and the fungus: A. niger. Surprisingly, the ligand and its palladium derivative showed only antibacterial activity towards S. aureus. The results indicated high antibacterial activities (inhibition zone: 26 mm for HL and 30 mm for Pd(HL)) and were found to be comparable with that of doxycycline standard (inhibition zone: 32 mm). Accordingly, it could be concluded that the Pd species improved the antibacterial activity relative to ligand. The high activity of the complex can be explained by the cell permeability concept and/or Tweedy’s chelation theory [44,45,46]. According to the cell permeability concept, the metal ions can hardly pass through the membrane surrounding the cell due to the high polarity of such ions. On chelation, the polarity of Pd(II) ions can be reduced to a greater extent as a result of the overlap of the ligand and metal ion orbitals leading to a partial sharing of the positive charge of the metal ion with donor groups. Furthermore, the lipophilicity of complex is improved as the π-electron delocalization over the whole chelating ring increases. Subsequently, the penetration of the complex into lipid membranes will be enhanced and then blocking the Pd(II) binding sites in the enzymes of microorganisms.
Molecular docking is a useful tool to understand the interaction between the synthesized compounds and biological targets. Analysis of the docking data is used to predict the conformational changes associated with the amino acid residues at the binding position to accommodate the docked hydrophobic inhibitors. The HL ligand and its zinc complex were subjected to molecular docking studies using Auto Dock Vina 4 to understand the compound-DNA and compound-protein interactions and to explore the potential binding mode and energy. The docked ligands conformations were rated according to the binding energy, hydrogen bonding, and hydrophobic interactions between the two compounds (HL and Zn(L)) with either the BDNA (PDB ID: 1ZEW) or the protein (PDB ID: 4MZI). The docking studies predict the way by which the docked compounds principally fit in the DNA or the protein minor grooves and how to involve of the hydrophobic, ionic, and hydrogen bonding interactions with the nitrogen bases. The binding interactions of HL and Zn(L) with the DNA and protein are displayed in Figures 4 and 5. The two derivatives showed very good binding scores, which indicate high binding affinity between the receptors (DNA and protein) and ligand (HL and Zn(L)) molecules. These values display their high efficiency as bioactive compounds (−9.1 and −6.9 kcal/mol for HL, and −8.9 and −7.3 kcal/mol for Zn(L), resp.). In case of HL, the binding interaction with DNA comes from either hydrophobic interaction between the amino acid residues or hydrogen bonding with the DG 17 and DA 16 regions and the hydroxyl group of the ligand. On the other hand, the binding interaction of HL and protein comes from hydrophobic interaction and hydrogen bonds formed between HIS 193, HIS 214, and VAL 253 parts of protein. The Zn(L) derivative also showed similar interaction with both DNA and protein. Interestingly, the interaction of Zn(L) with DNA is comparable to that of HL due to the presence water OH groups instead of the hydroxyl group of ligand (interaction occurred between the nitrogen and water OH of Zn(L) with the DG 17, DG 19, and DA 6 regions). In addition, more binding interaction of Zn(L) with other sites of the protein were observed (VAL 143, ILE 232, ASN 200, ALA 159).
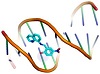
Figure 4: 3D representation of HL interaction with: DNA ((a) cartoon; (b) mesh) and protein ((c) cartoon; (d) mesh).
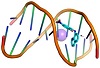
Figure 5: 3D representation of zinc complex interaction with: DNA ((a) cartoon; (b) mesh) and protein ((c) cartoon; (d) mesh).
The fourth author is the Editor-in-Chief of this journal; accordingly, Dr. S.M. El-Medani and the coauthors were blind to the details of the review. The authors have no other conflicts of interest.
Copyright © 2020 Ramadan M. Ramadan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.